14.8.1. steps.API_1.model
Namespace for objects of species, reactions and diffusions, as well as their containers.
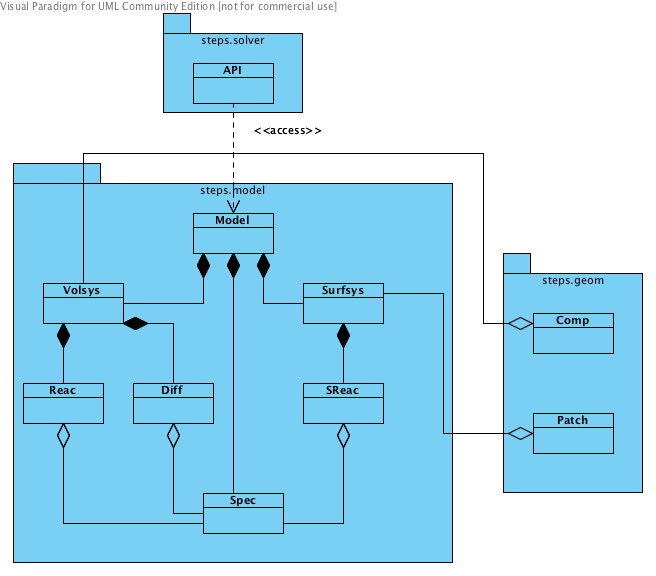
Class diagram for steps.API_1.model namespace.
- class Model[source]
Top-level container for the objects in a kinetic model.
Construction:
m = steps.model.Model()
Create a model container object.
Arguments: None
- delSpec(id)
Remove the steps.model.Spec species object with identifier string spec_id (if defined) from the model.
Syntax:
delSpec(spec_id)
Arguments: string spec_id
Return: None
- delSurfsys(id)
Remove the steps.model.Surfsys surface system object with identifier string ssys_id (if defined) from the model.
Syntax:
delSurfsys(ssys_id)
Arguments: string ssys_id
Return: None
- delVolsys(id)
Remove the steps.model.Volsys volume system object with identifier string vsys_id (if defined) from the model.
Syntax:
delVolsys(vsys_id)
Arguments: string vsys_id
Return: None
- getAllChans()
Returns a list of steps.model.Chan object references of all channels in the model.
Syntax:
getAllChans()
Arguments: None
Return: list<steps.model.Chan>
- getAllLinkSpecs()
- getAllRafts()
- getAllRaftsyss()
- getAllSpecs()
Returns a list of steps.model.Spec object references of all species in the model.
Syntax:
getAllSpecs()
Arguments: None
Return: list<steps.model.Spec>
- getAllSurfsyss()
Returns a list of steps.model.Surfsys object references of all surface systems in the model.
Syntax:
getAllSurfsyss()
Arguments: None
Return: list<steps.model.Surfsys>
- getAllVesSurfsyss()
- getAllVesicles()
- getAllVolsyss()
Returns a list of steps.model.Volsys object references of all volume systems in the model.
Syntax:
getAllVolsyss()
Arguments: None
Return: list<steps.model.Volsys>
- getChan(id)
Returns a reference to the steps.model.Chan channel object with identifier string chan_id (if defined).
Syntax:
getSpec(chan_id)
Arguments: string chan_id
Return: steps.model.Chan
- getLinkSpec(id)
- getRaft(id)
- getRaftsys(id)
- getSpec(id)
Returns a reference to the steps.model.Spec species object with identifier string spec_id (if defined).
Syntax:
getSpec(spec_id)
Arguments: string spec_id
Return: steps.model.Spec
- getSurfsys(id)
Returns a reference to the steps.model.Surfsys surface system object with identifier string ssys_id (if defined).
Syntax:
getSurfsys(ssys_id)
Arguments: string ssys_id
Return: steps.model.Surfsys
- getVesSurfsys(id)
- getVesicle(id)
- getVolsys(id)
Returns a reference to the steps.model.Volsys volume system object with identifier string vsys_id (if defined).
Syntax:
getVolsys(vsys_id)
Arguments: string vsys_id
Return: steps.model.Volsys
- this
An identification of the underlying cpp object
- class Spec(id, model, valence=0)[source]
A chemical species which can be a reactant and/or product in reaction stoichiometry and/or associated with a diffusion rule, or other transport mechanisms.
Construction:
s = steps.model.Spec(id, mdl)
Create a species object with identifier string id and assign the object mdl as its parent model.
Arguments: string id steps.model.Model mdl int valence (default=0)
- getID()
Get the identifier string of the species.
Syntax:
getID()
Arguments: None
Return: string
- getModel()
Returns a reference to the parent steps.model.Model container object.
Syntax:
getModel()
Arguments: None
Return: steps.model.Model
Attribute: model
- getValence()
Returns the valence of the species.
Syntax:
getValence()
Arguments: None
Return: int
- property id
Identifier string of the species.
- property model
Reference to parent model.
- setID(id)
Set the identifier string of the species.
Syntax:
setID(name)
Arguments: string name
Return: None
- setValence(valence)
Set the valence of the species.
Syntax:
setValence(valence)
Arguments: int valence
Return: None
- this
An identification of the underlying cpp object
- class Volsys(id, model)[source]
A container that groups reactions and diffusion rules involving Species located in a volume.
Construction:
v = steps.model.Volsys(id, mdl)
Construct a volume system object with identifier string id and assign the object mdl as its parent model.
Arguments: string id steps.model.Model mdl
- delDiff(id)
Remove the steps.model.Diff diffusion-rule object with identifier diff_id from the volume system.
Syntax:
delDiff(diff_id)
Arguments: string diff_id
Return: None
- delReac(id)
Remove the steps.model.Reac reaction-rule object with identifier reac_id (if defined) from the volume system.
Syntax:
delReac(reac_id)
Arguments: string reac_id
Return: None
- getAllComplexReacs()
Returns a list of references to all steps.model.ComplexReac complex reaction objects defined in the volume system.
Syntax:
getAllComplexReacs()
Arguments: None
Return: list<steps.model.ComplexReac>
- getAllDiffs()
Returns a list of references to all steps.model.Diff diffusion-rule objects defined in the volume system.
Syntax:
getAllDiffs()
Arguments: None
Return: list<steps.model.Diff>
- getAllReacs()
Returns a list of references to all steps.model.Reac objects in this volume system; that is all reaction rules belonging to this volume system. No duplicate member is included.
Syntax:
getAllReacs()
Arguments: None
Return: list<steps.model.Reac>
- getAllSpecs()
Returns a list of references to all steps.model.Spec objects in this volume system; that is all reactants, products or diffusing species in the reaction and diffusion rules belonging to this volume system. No duplicate member is included.
Syntax:
getAllSpecs()
Arguments: None
Return: list<steps.model.Spec>
- getAllVesBinds()
Returns a list of references to all steps.model.VesBind vesicle binding objects defined in the volume system.
Syntax:
getAllVesBinds()
Arguments: None
Return: list<steps.model.VesBind>
- getAllVesUnbinds()
Returns a list of references to all steps.model.VesUnbind vesicle unbinding objects defined in the volume system.
Syntax:
getAllVesUnbinds()
Arguments: None
Return: list<steps.model.VesUnbind>
- getDiff(id)
Returns a reference to the steps.model.Diff diffusion-rule object with identifier diff_id (if defined in the volume system).
Syntax:
getDiff(diff_id)
Arguments: string diff_id
Return: steps.model.Diff
- getID()
Get the identifier string of the volume system.
Syntax:
getID()
Arguments: None
Return: string
- getModel()
Returns a reference to the parent steps.model.Model container object.
Syntax:
getModel()
Arguments: None
Return: steps.model.Model
- getReac(id)
Returns a reference to the steps.model.Reac reaction-rule object with identifier string reac_id (if defined in the volume system).
Syntax:
getReac(reac_id)
Arguments: string reac_id
Return: steps.model.Reac
- getVesBind(id)
Returns a reference to the steps.model.VesBind vesicle binding object with identifier vesbind_id (if defined in the volume system).
Syntax:
getVesBind(vesbind_id)
Arguments: string vesbind_id
Return: steps.model.VesBind
- getVesUnbind(id)
Returns a reference to the steps.model.VesUnbind vesicle unbinding object with identifier vesunbind_id (if defined in the volume system).
Syntax:
getVesUnbind(vesunbind_id)
Arguments: string vesunbind_id
Return: steps.model.VesUnbind
- property id
Identifier string of the volume system.
- property model
Reference to parent model.
- setID(newid)
Set the identifier string of the volume system.
Syntax:
setID(name)
Arguments: string name
Return: None
- this
An identification of the underlying cpp object
- class Surfsys(id, model)[source]
A container that groups reactions, diffusion rules and other transport mechanisms involving a Species located in a membrane.
Construction:
s = steps.model.Surfsys(id, mdl)
Construct a surface system object with identifier string id and assign the object mdl as its parent model.
Arguments: string id steps.model.Model mdl
- delDiff(id)
Remove the steps.model.Diff diffusion-rule object with identifier diff_id from the surface system.
Syntax:
delDiff(diff_id)
Arguments: string diff_id
Return: None
- delGHKcurr(id)
Remove the steps.model.GHKcurr ghk current object with identifier id from the surface system.
Syntax:
delGHKcurr(id)
Arguments: string id
Return: None
- delOhmicCurr(id)
Remove the steps.model.OhmicCurr ohmic current object with identifier id from the surface system.
Syntax:
delOhmicCurr(id)
Arguments: string id
Return: None
- delSReac(id)
Remove the steps.model.SReac surface-reaction object with identifier id from the surface system.
Syntax:
delSReac(id)
Arguments: string id
Return: None
- delVDepSReac(id)
Remove the steps.model.VDepSReac voltage-dependent surface reaction object with identifier id from the surface system.
Syntax:
delVDepSReac(id)
Arguments: string id
Return: None
- getAllComplexSReacs()
Returns a list of references to all steps.model.ComplexSReac complex surface-reaction objects defined in the surface system.
Syntax:
getAllComplexSReacs()
Arguments: None
Return: list<steps.model.ComplexSReac>
- getAllDiffs()
Returns a list of references to all steps.model.Diff diffusion-rule objects defined in the surface system.
Syntax:
getAllDiffs()
Arguments: None
Return: list<steps.model.Diff>
- getAllEndocytosis()
Returns a list of references to all steps.model.Endocytosis endocytosis objects defined in the surface system.
Syntax:
getAllEndocytosis()
Arguments: None
Return: list<steps.model.Endocytosis>
- getAllGHKcurrs()
Returns a list of references to all steps.model.GHKcurr ghk current objects defined in the surface system.
Syntax:
getAllGHKcurrs()
Arguments: None
Return: list<steps.model.GHKcurrs>
- getAllOhmicCurrs()
Returns a list of references to all steps.model.OhmicCurr ohmic current objects defined in the surface system.
Syntax:
getAllOhmicCurrs()
Arguments: None
Return: list<steps.model.OhmicCurr>
- getAllRaftGens()
Returns a list of references to all steps.model.RaftGen raft generation objects defined in the surface system.
Syntax:
getAllRaftGens()
Arguments: None
Return: list<steps.model.RaftGen>
- getAllSReacs()
Returns a list of references to all steps.model.SReac surface-reaction objects defined in the surface system.
Syntax:
getAllSReacs()
Arguments: None
Return: list<steps.model.SReac>
- getAllSpecs()
Returns a list of references to all steps.model.Spec species objects included in the surface system; that is all reactants, products and channel states that appear in the interactions belonging to this surface system, regardless of where they appear in the geometry (i.e. includes all species in referenced patches and compartments). No duplicate member is included.
Syntax:
getAllSpecs()
Arguments: None
Return: list<steps.model.Spec>
- getAllVDepSReacs()
Returns a list of references to all steps.model.VDepSReac voltage-dependent surface reaction objects defined in the surface system.
Syntax:
getAllVDepSReacs()
Arguments: None
Return: list<steps.model.VDepSReac>
- getDiff(id)
Returns a reference to the steps.model.Diff diffusion-rule object with identifier diff_id (if defined in the surface system).
Syntax:
getDiff(diff_id)
Arguments: string diff_id
Return: steps.model.Diff
- getEndocytosis(id)
Returns a reference to the steps.model.Endocytosis endocytosis object with identifier id (if defined in the surface system).
Syntax:
getEndocytosis(id)
Arguments: string id
Return: steps.model.Endocytosis
- getGHKcurr(id)
Returns a reference to the steps.model.GHKcurr ghk current object with identifier id (if defined in the surface system).
Syntax:
getGHKcurr(id)
Arguments: string id
Return: steps.model.GHKcurr
- getID()
Get the identifier string of the surface system.
Syntax:
getID()
Arguments: None
Return: string
- getModel()
Returns a reference to the parent steps.model.Model container object.
Syntax:
getModel()
Arguments: None
Return: steps.model.Model
- getOhmicCurr(id)
Returns a reference to the steps.model.OhmicCurr ohmic current object with identifier id (if defined in the surface system).
Syntax:
getOhmicCurr(id)
Arguments: string id
Return: steps.model.OhmicCurr
- getRaftGen(id)
Returns a reference to the steps.model.RaftGen raft generation object with identifier id (if defined in the surface system).
Syntax:
getRaftGen(id)
Arguments: string id
Return: steps.model.RaftGen
- getSReac(id)
Returns a reference to the steps.model.SReac surface-reaction object with identifier sreac_id (if defined in the surface system).
Syntax:
getSReac(id)
Arguments: string id
Return: steps.model.SReac
- getVDepSReac(id)
Returns a reference to the steps.model.VDepSReac voltage-dependent surface reaction object with identifier id (if defined in the surface system).
Syntax:
getVDepSReac(id)
Arguments: string id
Return: steps.model.VDepSReac
- property id
Identifier string of the surface system.
- property model
Reference to parent model.
- setID(id)
Set the identifier string of the surface system.
Syntax:
setID(name)
Arguments: string name
Return: None
- this
An identification of the underlying cpp object
- class Reac(id, volsys, lhs=[], rhs=[], kcst=0)[source]
A kinetic reaction rule in a volume.
The reaction rule is specified by:
Species on the left hand side of the reaction: the reactants (lhs).
Species on the right hand side of the reaction: the products (rhs).
Rate constant for the reaction, supplied in s.i. units (kcst).
Construction:
reac = steps.model.Reac(id, volsys, lhs = [ ], rhs = [ ], kcst = 0.0)
Construct a reaction rule object with identifier string id and assign volsys as the parent volume system. A list of left hand side reactants may be assigned with lhs, whilst a list of right hand side products may be assigned with rhs, the kinetic reaction rate constant is set by kcst.
Arguments: string id steps.model.Volsys volsys list(steps.model.Spec) lhs (default = [ ]) list(steps.model.Spec) rhs (default = [ ]) float kcst (default = 0.0)
- getAllSpecs()
Returns a list of references to all steps.model.Spec species objects in the reaction; that is all reactants and products. No duplicate member is included.
Syntax:
getAllSpecs()
Arguments: None
Return: list<steps.model.Spec>
- getID()
Get the identifier string of the reaction rule.
Syntax:
getID()
Arguments: None
Return: string
- getKcst()
Get the kinetic reaction rate constant, in s.i. units, where the actual units depend on the order of the reaction.
Syntax:
getKcst()
Arguments: None
Return: float
- getLHS()
Get a list of references to steps.model.Spec species objects on the left hand side of the reaction: the reactants.
Syntax:
getLHS()
Arguments: None
Return: list<steps.model.Spec>
- getModel()
Returns a reference to the parent steps.model.Model container object.
Syntax:
getModel()
Arguments: None
Return: steps.model.Model
- getOrder()
Returns the order of this reaction.
Syntax:
getOrder()
Arguments: None
Return: int
- getRHS()
Get a list of references to steps.model.Spec species objects on the right hand side of the reaction: the reactants.
Syntax:
getRHS()
Arguments: None
Return: list<steps.model.Spec>
- getVolsys()
Returns a reference to the parent steps.model.Volsys volume system object.
Syntax:
getVolsys()
Arguments: None
Return: steps.model.Volsys
- property id
Identifier string of the reaction.
- property kcst
Reaction constant.
- property lhs
Left hand side reactants.
- property model
Reference to parent model.
- property order
Order of the reaction.
- property rhs
Right hand side reactants.
- setID(id)
Set the identifier string of the reaction rule.
Syntax:
setID(name)
Arguments: string name
Return: None
- setKcst(kcst)
Set the kinetic reaction rate constant, in s.i. units, where the actual units depend on the order of the reaction.
Syntax:
setKcst(kcst)
Arguments: float kcst
Return: None
- setLHS(lhs)
Set a list of references to steps.model.Spec species objects on the left hand side of the reaction: the reactants.
Syntax:
setLHS(lhs)
Arguments: list<steps.model.Spec> lhs
Return: None
- setRHS(rhs)
Set a list of references to steps.model.Spec species objects on the right hand side of the reaction: the reactants.
Syntax:
setRHS(rhs)
Arguments: list<steps.model.Spec> rhs
Return: None
- this
An identification of the underlying cpp object
- property volsys
Reference to parent volume system.
- class SReac(id, surfsys, olhs=[], ilhs=[], slhs=[], irhs=[], srhs=[], orhs=[], kcst=0)[source]
A reaction rule where at least one reactant or product is embedded in a surface.
In a surface reaction, the species can be classified as:
- Reactants on the left hand side of the reaction:
Species in the ‘outer’ compartment (olhs)
or Species in the ‘inner’ compartment (ilhs)
Species on the surface (slhs)
- Products on right hand side of the reaction:
Species in the ‘outer’ compartment (orhs)
Species in the ‘inner’ compartment (irhs)
Species on the surface (srhs)
The reaction rate is defined by kcst, supplied in s.i. units.
Construction:
sreac = steps.model.SReac(id, surfsys, ilhs = [ ], olhs = [ ], slhs = [ ], irhs = [ ], orhs = [ ], srhs = [ ], kcst = 0.0)
Construct a surface reaction rule object with identifier string id and assign surfsys as the parent surface system. A list of left hand reactants are assigned with ilhs, olhs and slhs (default for each is an empty list). A list of right hand side products are assigned with irhs, orhs and srhs (default for each is an empty list). The kinetic reaction rate constant is set with kcst.
Arguments: string id steps.model.Surfsys surfsys list(steps.model.Spec) ilhs (default = [ ]) list(steps.model.Spec) olhs (default = [ ]) list(steps.model.Spec) slhs (default = [ ]) list(steps.model.Spec) irhs (default = [ ]) list(steps.model.Spec) orhs (default = [ ]) list(steps.model.Spec) srhs (default = [ ]) float kcst (default = 0.0)
- getAllSpecs()
Returns a list of references to all steps.model.Spec species objects in the surface reaction; that is all reactants and products. No duplicate member is included.
Syntax:
getAllSpecs()
Arguments: None
Return: list<steps.model.Spec>
- getID()
Get the identifier string of the surface reaction rule.
Syntax:
getID()
Arguments: None
Return: string
- getILHS()
Get a list of references to steps.model.Spec species objects; the left hand side inner volume reactants.
Syntax:
getILHS()
Arguments: None
Return: list<steps.model.Spec>
- getIRHS()
Get a list of references to steps.model.Spec species objects; the right hand side inner volume reactants.
Syntax:
getIRHS()
Arguments: None
Return: list<steps.model.Spec>
- getKcst()
Get the kinetic reaction rate constant, in s.i. units, where the actual units depend on the order of the surface reaction.
Syntax:
getKcst()
Arguments: None
Return: float
- getModel()
Returns a reference to the parent steps.model.Model container object of parent surface system object.
Syntax:
getModel()
Arguments: None
Return: steps.model.Model
- getOLHS()
Get a list of references to steps.model.Spec species objects; the left hand side outer volume reactants.
Syntax:
getOLHS()
Arguments: None
Return: list<steps.model.Spec>
- getORHS()
Get a list of references to steps.model.Spec species objects; the right hand side outer volume reactants.
Syntax:
getORHS()
Arguments: None
Return: list<steps.model.Spec>
- getOrder()
Returns the order of this surface reaction.
Syntax:
getOrder()
Arguments: None
Return: int
- getSLHS()
Get a list of references to steps.model.Spec species objects; the left hand side surface reactants.
Syntax:
getSLHS()
Arguments: None
Return: list<steps.model.Spec>
- getSRHS()
Get a list of references to steps.model.Spec species objects; the right hand side surface reactants.
Syntax:
getSRHS()
Arguments: None
Return: list<steps.model.Spec>
- getSurfsys()
Returns a reference to the parent steps.model.Surfsys surface system object.
Syntax:
getSurfsys()
Arguments: None
Return: steps.model.Surfsys
- property id
Identifier string of the surface reaction.
- property ilhs
Left hand side reactants in inner compartment.
- property irhs
Right hand side reactants in inner compartment.
- property kcst
Reaction constant.
- property model
Reference to parent model.
- property olhs
Left hand side reactants in outer compartment.
- property order
Order of the reaction.
- property orhs
Right hand side reactants in outer compartment.
- setID(id)
Set the identifier string of the surface reaction rule.
Syntax:
setID(name)
Arguments: string name
Return: None
- setILHS(ilhs)
Set a list of references to steps.model.Spec species objects; the left hand side inner volume reactants.
Syntax:
setILHS(ilhs)
Arguments: list<steps.model.Spec> ilhs
Return: None
- setIRHS(irhs)
Set a list of references to steps.model.Spec species objects; the right hand side inner volume reactants.
Syntax:
setIRHS(irhs)
Arguments: list<steps.model.Spec> irhs
Return: None
- setKcst(kcst)
Set the kinetic reaction rate constant, in s.i. units, where the actual units depend on the order of the surface reaction.
Syntax:
setKcst(kcst)
Arguments: float kcst
Return: None
- setOLHS(olhs)
Set a list of references to steps.model.Spec species objects; the left hand side outer volume reactants.
Syntax:
setOLHS(olhs)
Arguments: list<steps.model.Spec) olhs
Return: None
- setORHS(orhs)
Get a list of references to steps.model.Spec species objects; the right hand side outer volume reactants.
Syntax:
setORHS(orhs)
Arguments: list<steps.model.Spec> orhs
Return: None
- setSLHS(slhs)
Set a list of references to steps.model.Spec species objects; the left hand side surface reactants.
Syntax:
setSLHS(slhs)
Arguments: list<steps.model.Spec> slhs
Return: None
- setSRHS(srhs)
Set a list of references to steps.model.Spec species objects; the right hand side surface reactants.
Syntax:
setSRHS(srhs)
Arguments: list<steps.model.Spec> srhs
Return: None
- property slhs
Left hand side reactants on surface.
- property surfsys
Reference to parent surface system.
- this
An identification of the underlying cpp object
- class Diff(id, volsys_or_surfsys, lig, dcst=0)[source]
A diffusion rule for a chemical species in a volume or on a surface.
A diffusion rule is described by:
Species to which the diffusion rule applies (lig).
Diffusion constant (dcst) specified in s.i. units.
Construction:
diff = steps.model.Diff(id, volsys, lig, dcst = 0.0)
Construct a diffusion rule object with identifier string id applied to species lig and assign volsys as the parent volume system. Diffusion constant is set by dcst.
Arguments: string id steps.model.Volsys volsys steps.model.Spec lig float dcst (default = 0.0)
Construction:
diff = steps.model.Diff(id, surfsys, lig, dcst = 0.0)
Construct a diffusion rule object with identifier string id applied to species lig and assign surfsys as the parent surface system. Diffusion constant is set by dcst.
Arguments: string id steps.model.Surfsys surfsys steps.model.Spec lig float dcst (default = 0.0)
- property dcst
Diffusion constant.
- getAllSpecs()
Return a reference to the steps.model.Spec species object to which this diffusion rule is applied as a list of length 1.
Syntax:
getAllSpecs()
Arguments: None
Return: list<steps.model.Spec, length=1>
- getDcst()
Get the diffusion constant for the diffusion rule, in s.i. units.
Syntax:
getDcst()
Arguments: None
Return: float
- getID()
Get the identifier string of the diffusion rule.
Syntax:
getID()
Arguments: None
Return: string
- getLig()
get a reference to the steps.model.Spec species object to which this diffusion rule is applied.
Syntax:
getLig()
Arguments: None
Return: steps.model.Spec
- getModel()
Returns a reference to the parent steps.model.Model container object.
Syntax:
getModel()
Arguments: None
Return: steps.model.Model
- getSurfsys()
Returns a reference to the parent steps.model.Surfsys surface system object if surface diffusion object.
Syntax:
getSurfys()
Arguments: None
Return: steps.model.Surfsys
- getVolsys()
Returns a reference to the parent steps.model.Volsys volume system object if a volume diffusion object.
Syntax:
getVolsys()
Arguments: None
Return: steps.model.Volsys
- property id
Identifier string of the diffusion.
- property lig
Reference to diffusion species.
- property model
Reference to parent model.
- setDcst(dcst)
Set the diffusion constant for the diffusion rule, in s.i. units.
Syntax:
setDcst(dcst)
Arguments: float dcst
Return: None
- setID(id)
Set the identifier string of the diffusion rule.
Syntax:
setID(name)
Arguments: string name
Return: None
- setLig(lig)
Set a reference to the steps.model.Spec species object to which this diffusion rule is applied.
Syntax:
setLig(lig)
Arguments: steps.model.Spec lig
Return: None
- property surfsys
Reference to parent surface system.
- this
An identification of the underlying cpp object
- property volsys
Reference to parent volume system.
- class Chan(id='')[source]
A channel object which groups a set of channel states. Whilst not involved directly in reactions, currents, transitions etc it provides necessary grouping for channel states.
Construction:
chan = steps.model.Chan(id, mdl)
Create a channel object with identifier string id and assign the object mdl as its parent model.
Arguments: string id steps.model.Model mdl
- getAllChanStates()
Returns a list of steps.model.Chanstate object references of all channel states in the channel.
Syntax:
getAllChanStates()
Arguments: None
Return: list<steps.model.ChanState>
- getChanState(id)
Returns a reference to channel state of the channel with string identifier id.
Syntax:
getChanState(id)
Arguments: string id
Return: steps.model.Chanstate
- getID()
Get the identifier string of the channel.
Syntax:
getID()
Arguments: None
Return: string
- getModel()
Returns a reference to the parent steps.model.Model container object.
Syntax:
getModel()
Arguments: None
Return: steps.model.Model
Attribute: model
- property id
Identifier string of the channel.
- property model
Refernece to parent model.
- setID(id)
Set the identifier string of the channel.
Syntax:
setID(name)
Arguments: string name
Return: None
- this
An identification of the underlying cpp object
- class ChanState(id, model, chan)[source]
A channel state object which may be involved in channel events such as currents, voltage-dependent transitions and any other steps.model.Spec events, such as surface reactions and diffusion.
Construction:
chanstate = steps.model.ChanState(id, mdl, chan)
Create a channel state object with identifier string id, assign the object mdl as its parent model. This object describes one of the possible states of channel object chan.
Arguments: string id steps.model.Model mdl steps.model.Chan chan
- property chan
Reference to parent channel.
- getChan()
Returns a reference to the parent steps.model.Chan container object.
Syntax:
getChan()
Arguments: None
Return: steps.model.Chan
- getID()
Get the identifier string of the species.
Syntax:
getID()
Arguments: None
Return: string
- getModel()
Returns a reference to the parent steps.model.Model container object.
Syntax:
getModel()
Arguments: None
Return: steps.model.Model
Attribute: model
- getValence()
Returns the valence of the species.
Syntax:
getValence()
Arguments: None
Return: int
- property id
Identifier string of the species.
- property model
Reference to parent model.
- setID(id)
Set the identifier string of the channel state.
Syntax:
setID(name)
Arguments: string name
Return: None
- setValence(valence)
Set the valence of the species.
Syntax:
setValence(valence)
Arguments: int valence
Return: None
- this
An identification of the underlying cpp object
- class GHKcurr(id, surfsys, chanstate, ion, computeflux=True, virtual_oconc=- 1.0, vshift=0.0)[source]
A current object based on the Goldman-Hodgkin-Katz flux equation, that describes a current through a channel in a particular permeable state.
Each GHK current in the simulation is solved within the SSA with a rate determined from the simulation state, i.e. membrane potential, ‘outer’ and ‘inner’ concentration of the ion and temperature (which is fixed), and constant parameters ion valence and permeability. For a permeabilty to be found the user must supply information from a channel measurement:
the single-channel conductance
the potential
the temperature (this may be different from the STEPS simulation temperature)
the ‘outer’ concentration of the ion
the ‘inner’ concentration of the ion
This information must be set with the member function setPInfo()
A GHK current involves, optionally, a real transfer of ions between compartments separated by a membrane in the STEPS simulation. It is important that these ions implement diffusion if compartments are not well-mixed.
Construction:
ghkcurr = steps.model.GHKcurr(id, surfsys, chanstate, ion, computeflux=True)
Construct a ghk current object with identifier string id and assign surfsys as the parent surface system. Assign to channel state chanstate, set the species that describes the current with ion- this species object must have a valence specified. If computeflux flag is set to True then the current will result in movement of ions between compartments, if False the current will be calculated but will not correspond to a real ion flux. A ‘virtual outer concentration’ can be specified so that the outer compartment does not have to be explicitly simulated (if it retains default negative value then the outer concentration of ion must be simulated).
Arguments: string id steps.model.Surfsys surfsys steps.model.ChanState chanstate steps.model.Spec ion bool computeflux float virtual_oconc (default = -1.0)
NOTE: function setP or setPInfo must be called on the object before creating simulation object.
- property chanState
_py_GHKcurr.getChanState(self)
Returns a reference to the steps.model.ChanState channel state object.
Syntax:
getChanState()
Arguments: None
Return: steps.model.ChanState
- getChanState()
Returns a reference to the steps.model.ChanState channel state object.
Syntax:
getChanState()
Arguments: None
Return: steps.model.ChanState
- getID()
Get the identifier string of the ghk current.
Syntax:
getID()
Arguments: None
Return: string
- getIon()
Returns a reference to steps.model.Spec object- the ion of this ghk current.
Syntax:
getIon()
Arguments: None
Return: steps.model.Spec
- getModel()
Returns a reference to the parent steps.model.Model container object of parent surface system object.
Syntax:
getModel()
Arguments: None
Return: steps.model.Model
- getP()
Get the single-channel permeability (units: cubic meters / second).
Syntax:
getP()
Arguments: None
Return: float
- getSurfsys()
Returns a reference to the parent steps.model.Surfsys surface system object.
Syntax:
getSurfsys()
Arguments: None
Return: steps.model.Surfsys
- property id
Identifier string of the ghk current.
- property ion
The current ion.
- property model
Reference to parent model.
- setChanState(chanstate)
Set the channel state for this ghk current.
Syntax:
setChanState(chanstate)
Arguments: steps.model.ChanState chanstate
Return: None
- setID(id)
Set the identifier string of the ghk current.
Syntax:
setID(name)
Arguments: string name
Return: None
- setIon(ion)
Set the ion for this ghk current.
Syntax:
setIon(ion)
Arguments: steps.model.Spec ion
Return: None
- setP(p)
Set the single-channel permeability directly (units: cubic meters / second).
Syntax:
setP(p)
Arguments: float p (the single-channel permeability)
Return: None
- setPInfo(g, V, T, oconc, iconc)
Supply information from a channel measurement in order to find the permeability. A measured single-channel coonductance (in Siemens) should be supplied, along with the potential (in volts), temperature (in Kelvins), the ‘outer’ concentration and ‘inner’ concentration of ion (in molar units).
Syntax:
setPInfo(g, V, T, oconc, iconc)
Arguments: float g (the conductance) float V (the voltage) float T (the temperatur float oconc (the ‘outer’ concentration) float iconc (the ‘inner’ concentration)
Return: None
- property surfsys
Reference to parent surface system.
- this
An identification of the underlying cpp object
- class OhmicCurr(id, surfsys, chanstate, erev, g)[source]
An ohmic current object that describes an ohmic current through a channel in a particular conducting state.
An ohmic current object is a simple current based on a single, fixed value for single-channel conductance and a constant reversal potential. An ohmic current does not result in movement of ions between compartments, but simply contributes a continous current to the EField solver for every conducting channel on a membrane surface that contains the ohmic current.
Construction:
ohmiccurr = steps.model.OhmicCurr(id, surfsys, chanstate, erev, g)
Construct an ohmic current object with identifier string id and assign surfsys as the parent surface system. Assign to channel state chanstate, set the reversal potential to erev (in volts) and the single-channel conductance to g (in Siemens).
Arguments: string id steps.model.Surfsys surfsys steps.model.ChanState chanstate float erev float g
- property erev
The reversal potential (in volts).
- property g
The single-channel conductance (in Siemens).
- getChanState()
Returns a reference to the steps.model.ChanState channel state object.
Syntax:
getChanState()
Arguments: None
Return: steps.model.ChanState
- getERev()
Returns the reversal potential of this ohmic current in volts.
Syntax:
getERev()
Arguments: None
Return: float
- getG()
Returns the single-channel conductance for this ohmic current in Siemens.
Syntax:
getG()
Arguments: None
Return: float
- getID()
Get the identifier string of the ohmic current.
Syntax:
getID()
Arguments: None
Return: string
- getModel()
Returns a reference to the parent steps.model.Model container object of parent surface system object.
Syntax:
getModel()
Arguments: None
Return: steps.model.Model
- getSurfsys()
Returns a reference to the parent steps.model.Surfsys surface system object.
Syntax:
getSurfsys()
Arguments: None
Return: steps.model.Surfsys
- property id
Identifier string of the ohmic current.
- property model
Reference to parent model.
- setChanState(chanstate)
Set the channel state for this ohmic current.
Syntax:
setChanState(chanstate)
Arguments: steps.model.ChanState chanstate
Return: None
- setERev(erev)
Set the reveral potential for this ohmic current in volts.
Syntax:
setERev(erev)
Arguments: float erev
Return: None
- setG(g)
Set the single-channel conductance for this ohmic current in Siemens.
Syntax:
setG(g)
Arguments: float g
Return: None
- setID(id)
Set the identifier string of the ohmic current.
Syntax:
setID(name)
Arguments: string name
Return: None
- property surfsys
Reference to parent surface system.
- this
An identification of the underlying cpp object
- class VDepSReac(id, surfsys, **kwargs)[source]
A voltage-dependent reaction involving any species or channel state.
A voltage-dependent reaction is similar to a surface reaction, except the reaction parameter is voltage-dependent.
A function object must be passed to the constructor returning the reaction ‘constant’ as a function of voltage. This function is used to fill a table of reaction ‘constants’ at a range of voltage values (default: -150mV to 100mV, in steps of 0.1mV) These default values may be altered with optional argument ‘vrange’.
- getAllSpecs()
Returns a list of references to all steps.model.Spec species objects in the voltage-dependent reaction; that is all reactants and products. No duplicate member is included.
Syntax:
getAllSpecs()
Arguments: None
Return: list<steps.model.Spec>
- getID()
Get the identifier string of the voltage-dependent reaction.
Syntax:
getID()
Arguments: None
Return: string
- getILHS()
Get a list of references to steps.model.Spec species objects; the left hand side inner volume reactants.
Syntax:
getILHS()
Arguments: None
Return: list<steps.model.Spec>
- getIRHS()
Get a list of references to steps.model.Spec species objects; the right hand side inner volume reactants.
Syntax:
getIRHS()
Arguments: None
Return: list<steps.model.Spec>
- getK()
Return a list of reaction ‘constants’ in the default voltage range.
Syntax:
getK()
Arguments: None
Return: list<float>
- getModel()
Returns a reference to the parent steps.model.Model container object of parent surface system object.
Syntax:
getModel()
Arguments: None
Return: steps.model.Model
- getOLHS()
Get a list of references to steps.model.Spec species objects; the left hand side outer volume reactants.
Syntax:
getOLHS()
Arguments: None
Return: list<steps.model.Spec>
- getORHS()
Get a list of references to steps.model.Spec species objects; the right hand side outer volume reactants.
Syntax:
getORHS()
Arguments: None
Return: list<steps.model.Spec>
- getOrder()
Returns the order of this voltage-dependent reaction.
Syntax:
getOrder()
Arguments: None
Return: int
- getSLHS()
Get a list of references to steps.model.Spec species objects; the left hand side surface reactants.
Syntax:
getSLHS()
Arguments: None
Return: list<steps.model.Spec>
- getSRHS()
Get a list of references to steps.model.Spec species objects; the right hand side surface reactants.
Syntax:
getSRHS()
Arguments: None
Return: list<steps.model.Spec>
- getSurfsys()
Returns a reference to the parent steps.model.Surfsys surface system object.
Syntax:
getSurfsys()
Arguments: None
Return: steps.model.Surfsys
- property id
Identifier string of the voltage-dependent reaction.
- property ilhs
Left hand side reactants in inner compartment.
- property irhs
Right hand side reactants in inner compartment.
- property model
Reference to parent model.
- property olhs
left hand side reactants in outer compartment.
- property order
Order of the voltage-dependent reaction.
- property orhs
Right hand side reactants in outer compartment.
- setID(id)
Set the identifier string of the voltage-dependent reaction.
Syntax:
setID(name)
Arguments: string name
Return: None
- setILHS(ilhs)
Set a list of references to steps.model.Spec species objects; the left hand side inner volume reactants.
Syntax:
setILHS(ilhs)
Arguments: list<steps.model.Spec> ilhs
Return: None
- setIRHS(irhs)
Set a list of references to steps.model.Spec species objects; the right hand side inner volume reactants.
Syntax:
setIRHS(irhs)
Arguments: list<steps.model.Spec> irhs
Return: None
- setOLHS(olhs)
Set a list of references to steps.model.Spec species objects; the left hand side outer volume reactants.
Syntax:
setOLHS(olhs)
Arguments: list<steps.model.Spec> olhs
Return: None
- setORHS(orhs)
Get a list of references to steps.model.Spec species objects; the right hand side outer volume reactants.
Syntax:
setORHS(orhs)
Arguments: list<steps.model.Spec> orhs
Return: None
- setSLHS(slhs)
Set a list of references to steps.model.Spec species objects; the left hand side surface reactants.
Syntax:
setSLHS(slhs)
Arguments: list<steps.model.Spec> slhs
Return: None
- setSRHS(srhs)
Set a list of references to steps.model.Spec species objects; the right hand side surface reactants.
Syntax:
setSRHS(srhs)
Arguments: list<steps.model.Spec> srhs
Return: None
- property slhs
Left hand side reactants on surface.
- property srhs
Right hand side reactants on surface.
- property surfsys
Reference to parent surface system.
- this
An identification of the underlying cpp object